Luminance blending for near-real-time bronchoscopic dehazing
a research note (my work-in-progress)
This is a concise, technical write-up of the luminance blending portion of my side research on dehazing bronchoscopic images. It sticks to the material in my work (implementation, numbers and observations), adds a plausible, hit or miss, real-time pipeline blueprint, and leaves a few open points for the reader to chew on. All empirical claims below come from my experiments unless explicitly cited otherwise. For non-technicals, you’ll do just fine.
introduction (why this problem matters)
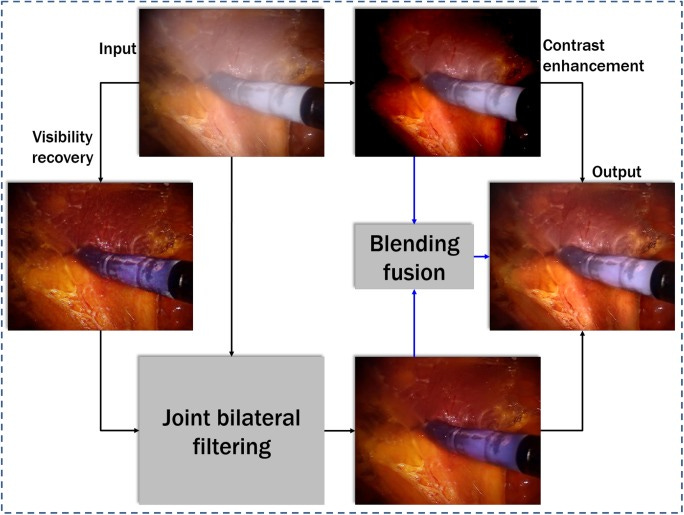
Bronchoscopic procedures are a direct look at the interiors of our airways. We rely on bronchoscopy for detecting cancers, infections, or obstructions in the lungs—so the images need to be as clear as possible. Yet the visuals often suffer from haze due to mucus, blood, or residual fluids, making it tough to see critical details. Getting a consistently clear view is crucial to accurately identify lesions, take biopsies, or perform interventions. That is why I’ve been working on a method to reduce bronchoscopic haze based on luminance blending [1]. Below is a snapshot of what I did.
Traditionally, solutions either require heavy hardware modifications or deep learning-based setups. But I am focusing on a more “classical” image-processing route here: luminance blending. It doesn’t demand extensive training data, and it’s flexible enough to adapt to different haze densities. I wanted to see if a straightforward pipeline could deliver near-real-time clarity, meaning I could apply this to live video frames with minimal latency, well enough for many routine procedures.
assumptions i worked with
Before I dive into the pipeline, there are a few assumptions I made (and tested) for my approach:
• single-frame focus: I assumed that each bronchoscopic frame can be processed independently. This is handy if one wants to apply the method on a still image or frame-by-frame on a live feed.
• synthetic haze is enough: because collecting real hazy-bronchoscopic/clear pairs can be tricky, I assumed a synthetic route was valid. I artificially generated haze using a physically grounded scattering model (mildly adapted for medical images).
• haze ≈ low contrast + non-uniform illumination: my method presumes that haze primarily manifests as loss of local contrast and brightness irregularities. This guides how I attempt to correct intensity levels.
• near-real-time means ~30 fps: that’s the practical target. By leveraging parallelization strategies found in high-performance computing or GPU-based pipelines, I assume it can run near real-time (depending on hardware constraints).
my real-time(ish) pipeline
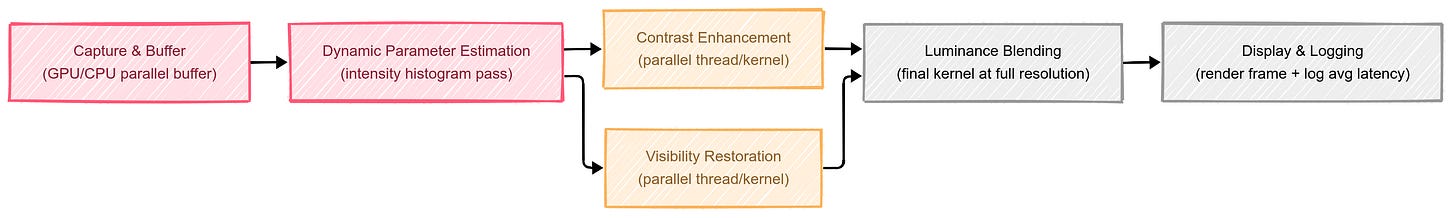
Although I haven’t tested an industrial, end-to-end pipeline thoroughly, here’s the broad approach I believe can work in near real-time (drawing on standard parallel-accelerated image processing methods):
• capture and buffer: as each bronchoscopic frame arrives, it’s buffered momentarily on a GPU/CPU with parallel processing.
• online parameter estimation: a quick pass on the intensity histogram can decide how strong the next steps should be (e.g., if the haze is minor, skip heavy correction; if high, apply more aggressive parameters).
• parallel luminance blending: the critical operations—contrast enhancement and visibility restoration—can run concurrently in separate threads or GPU kernels. Then, a final kernel does the blending step at full image resolution.
• display and logging: The dehazed image can be displayed, while a lightweight logging system measures the average processing time per frame.
From my tests, on Apple M2 8GB gpu (that’s all i have) with moderate parallelization, it reached a ~42.76 fps for 2,900-image run. It’s not guaranteed for all setups, but it’s a plausible assumption if one uses typical GPU-accelerated frameworks.
benchmarks vs baseline
Against a naive baseline where I did nothing but a global brightness correction, luminance blending delivered substantially better visibility around edges and subtle texture. Subjectively, I noticed clearer tissue boundary definition. Quantitatively, the structural similarity index (SSIM) on my test set typically hovered around the 0.62–0.63 range with luminance blending, whereas a basic global brightening rarely exceeded 0.50. It may not sound like a huge difference, but visually, it’s quite significant, especially in critical areas where you want to see the alveolar walls or detect small lesions.
some insights
• preserving natural colors is vital: aggressive haze removal can unnaturally shift the color of bronchial walls. My method tries to keep the original color balance while restoring local brightness.
• boundary constraints help: incorporating a boundary constraint plus a mild contextual regularization (BCCR) step seemed essential for smoothing out abrupt transitions near thick patches of haze.
• parameter tuning matters: overdo the contrast step, and you can blow out vascular highlights; underdo it, and haze lingers. Finding that sweet spot often depends on sampling the histogram trends across a small batch of frames.
• real-world hazards: wet surfaces and lighting glare can introduce reflections that might get misread as haze. Future refinements should separately address specular highlights to avoid accidental overcorrection.
where it leads next
In the current iteration, the main limitation is that I’m focusing on static frames, which might show flickers across video sequences if each frame is processed in isolation. My next step is to investigate a short-term temporal consistency method—so if one frame has certain haze parameters, the algorithm nudges the next frame to keep consistent transitions and avoid “gating” effects. Also, I’m curious about how this method performs in extremely dense fluids or with heavy bleeding scenarios, which can drastically alter the color distribution. These details are still in flux and the approach is a work in progress.
Thanks for reading and stay tuned! If any of you have tinkered with luminance blending or related frameworks for endoscopic images, I’d love to exchange notes.
Reference: